node2vec_online
The node2vec_online algorithm learns and updates temporal node embeddings on
the fly for tracking and measuring node similarity over time in graph streams.
The algorithm creates similar embeddings for two nodes (e.g. v and u) if there
is an option to reach one node from the other across edges that appeared
recently. In other words, the embedding of a node v should be more similar to
the embedding of node u if we can reach u by taking steps backward to node
v across edges that appeared before the previous one. These steps backward
from one node to the other form a temporal walk. It is temporal since it depends
on when the edge appeared in the graph.
To make two nodes more similar and to create these temporal walks, the Node2Vec
Online algorithm uses the StreamWalk updater and Word2Vec learner.
StreamWalk updater is a machine for sampling temporal walks. A sampling of the
walk is done in a backward fashion because we look only at the incoming edges of
the node. Since one node can have multiple incoming edges, when sampling a walk,
StreamWalk updater uses probabilities to determine which incoming edge of the
node it will take next, and that way leading to a new node. These probabilities
are computed after the edge arrives and before temporal walk sampling.
Probability represents a sum over all temporal walks z ending in node v
using edges arriving no later than the latest one of already sampled ones in the
temporal walk. When the algorithm decides which edge to take next for temporal
walk creation, it uses these computed weights (probabilities). Every time a new
edge appears in the graph, these probabilities are updated just for two nodes of
a new edge.
After walks sampling, Word2Vec learner uses these prepared temporal walks to
make node embeddings more similar using the gensim Word2Vec module. These
sampled walks are given as sentences to the gensim Word2Vec module, which then
optimizes for the similarity of the node embeddings in the walk with stochastic
gradient descent using a skip-gram model or continuous-bag-of-words (CBOW).
Embeddings capture the graph topology, relationships between nodes, and further relevant information. How the embeddings capture this inherent information of the graph is not fixed.
Capturing information in networks often shuttles between two kinds of similarities: homophily and structural equivalence. Under the homophily hypothesis, nodes that are highly interconnected and belong to similar network clusters or communities should be embedded closely together. In contrast, under the structural equivalence hypothesis, nodes that have similar structural roles in networks should be embedded closely together (e.g., nodes that act as hubs of their corresponding communities).
Currently, our implementation captures for homophily - nodes that are highly interconnected and belong to similar network clusters or communities.
1 Node embeddings in dynamic graphs, Ferenc Béres, Róbert Pálovics, Domokos Miklós Kelen and András A. Benczúr
| Trait | Value |
|---|---|
| Module type | module |
| Implementation | Python |
| Graph direction | directed |
| Edge weights | unweighted |
| Parallelism | sequential |
If this algorithm implementation is too slow for your use case, contact us and request a rewrite to C++ !
Procedures
If you want to execute this algorithm on graph projections, subgraphs or portions of the graph, be sure to check out the guide on How to run a MAGE module on subgraphs.
set_streamwalk_updater(half_life, max_length, beta, cutoff, sampled_walks, full_walks)
Input:
half_life: integer➡ half-life [seconds], used in the temporal walk probability calculationmax_length: integer➡ Maximum length of the sampled temporal random walksbeta: float➡ Damping factor for long pathscutoff: integer➡ Temporal cutoff in seconds to exclude very distant pastsampled_walks: integer➡ Number of sampled walks for each edge updatefull_walks: boolean➡ Return every node of the sampled walk for representation learning (full_walks=True) or only the endpoints of the walk (full_walks=False)
Output:
message: string➡ Whether parameters are set or they need to be reset
Usage:
CALL node2vec_online.set_streamwalk_updater(7200, 3, 0.9, 604800, 4, False);
set_word2vec_learner(embedding_dimension, learning_rate, skip_gram )
Input:
embedding_dimension: integer➡ Number of dimensions in the representation of the embedding vectorlearning_rate: float➡ Learning rateskip_gram: boolean➡ Whether to use skip-gram model (True) or continuous-bag-of-words (CBOW)negative_rate: integer➡ Negative rate for Gensim Word2Vec modelthreads: integer➡ Maximum number of threads for parallelization
Output:
message: string➡ Whether parameters are set or they need to be reset
Usage:
CALL node2vec_online.set_word2vec_learner(128, 0.01, True, 10, 1);
get()
Output:
node: mgp.Vertex➡ Node in the graph for which embedding existsembedding: mgp.List[mgp.Number]➡ Embedding for the given node
Usage:
CALL node2vec_online.get();
update(edges)
Input:
edges: mgp.List[mgp.Edge]➡ List of edges added to the graph. For those nodes onlynode2vec_onlinecalculates embeddings.
Usage:
There are a few options here. The first one is to create a trigger, so every time an edge is added to graph, the trigger calls a procedure and makes an update.
CREATE TRIGGER trigger ON --> CREATE BEFORE COMMIT
EXECUTE CALL node2vec_online.update(createdEdges) YIELD *;
The second option is to add all the edges and then call the algorithm with those edges:
MATCH (n)-[e]->(m)
WITH COLLECT(e) as edges
CALL node2vec_online.update(edges) YIELD *
WITH 1 as x
RETURN x;
reset()
Output:
message: string➡ Message that parameters are ready to be set again
Usage:
CALL node2vec_online.reset();
help()
Output:
name: string➡ Name of available functionsvalue: string➡ Documentation for every function
Usage:
CALL node2vec_online.help();
Example
- Step 1: Input graph
- Step 2: Set parameters and trigger
- Step 3: Load commands
- Step 4: Running command
- Step 5: Results
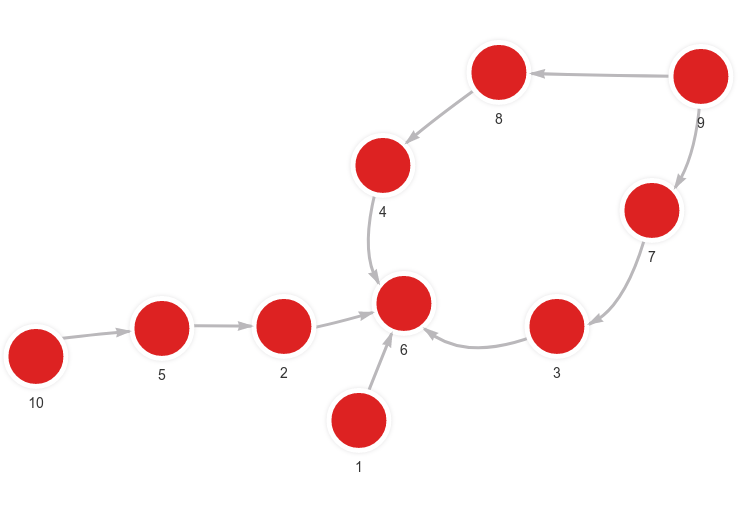
CALL node2vec_online.set_streamwalk_updater(7200, 2, 0.9, 604800, 2, True) YIELD *;
CALL node2vec_online.set_word2vec_learner(2, 0.01, True, 1, 1) YIELD *;
CREATE TRIGGER trigger ON --> CREATE BEFORE COMMIT
EXECUTE CALL node2vec_online.update(createdEdges) YIELD *;
MERGE (n:Node {id: 1}) MERGE (m:Node {id: 6}) CREATE (n)-[:RELATION]->(m);
MERGE (n:Node {id: 2}) MERGE (m:Node {id: 6}) CREATE (n)-[:RELATION]->(m);
MERGE (n:Node {id: 10}) MERGE (m:Node {id: 5}) CREATE (n)-[:RELATION]->(m);
MERGE (n:Node {id: 5}) MERGE (m:Node {id: 2}) CREATE (n)-[:RELATION]->(m);
MERGE (n:Node {id: 9}) MERGE (m:Node {id: 7}) CREATE (n)-[:RELATION]->(m);
MERGE (n:Node {id: 7}) MERGE (m:Node {id: 3}) CREATE (n)-[:RELATION]->(m);
MERGE (n:Node {id: 3}) MERGE (m:Node {id: 6}) CREATE (n)-[:RELATION]->(m);
MERGE (n:Node {id: 9}) MERGE (m:Node {id: 8}) CREATE (n)-[:RELATION]->(m);
MERGE (n:Node {id: 8}) MERGE (m:Node {id: 4}) CREATE (n)-[:RELATION]->(m);
MERGE (n:Node {id: 4}) MERGE (m:Node {id: 6}) CREATE (n)-[:RELATION]->(m);
CALL node2vec_online.get() YIELD node, embedding
RETURN node, embedding
ORDER BY node.id;
+-------------------------+-------------------------+
| node | embedding |
+-------------------------+-------------------------+
| (:Node {id: 1}) | [0.255167, 0.450464] |
| (:Node {id: 2}) | [-0.465147, -0.35584] |
| (:Node {id: 3}) | [-0.243008, -0.0908009] |
| (:Node {id: 4}) | [-0.414261, -0.472441] |
| (:Node {id: 5}) | [-0.250771, -0.188169] |
| (:Node {id: 6}) | [-0.0268114, 0.0118215] |
| (:Node {id: 7}) | [-0.226831, 0.327703] |
| (:Node {id: 8}) | [0.143829, 0.0495937] |
| (:Node {id: 9}) | [0.369025, -0.0766736] |
| (:Node {id: 10}) | [0.322944, 0.448649] |
+-------------------------+-------------------------+
