link_prediction_with_gnn
Link prediction can be defined as a problem where one wants to predict if there is a link between two nodes in the graph. It can be used for predicting missing or future links in the evolving graph. Using the notation G = (V, E) for a graph with nodes V and edges E and given two nodes v1 and v2, the link prediction algorithm tries to predict whether those two nodes will be connected, based on the node features and graph structure. Lately, graph neural networks have been often used for node-classification and link-prediction problems. They are extremely useful in numerous interdisciplinary fields of work where is important to incorporate domain-specific knowledge to capture more fine-grained relationships among the data. Such fields usually involve working with heterogeneous and large-scale graphs. GNNs iteratively update node representations by aggregating the representations of node neighbors and their representation from the previous iteration. Such properties make graph neural networks a great tool for various problems we in Memgraph encounter. If your graph is evolving in time, check TGN model that Memgraph engineers have already developed.
Blog Posts
The following blog posts explain how we tried to apply link prediction:
About the query module
In this module you can find support for the following interesting features:
- support for both homogeneous and heterogeneous graphs
- support for disconnected graphs
- its applicability to use it as a recommendation system
- a semi-inductive link prediction setup where a larger, updated graph is used for the inference
- an inductive link prediction setup in which training and inference graphs are different
- transductive graph splitting (training and validation sets)
- Graph attention layer aggregates information using an attention mechanism from the first-hop neighbourhood. Introduced by Velickovic et al.
- GraphSAGE layer extends the usability of graph neural networks to large-scale graphs. Introduced by Hamilton et al.
- mlp and dot predictors are used for combining node scores to edge scores
- ADAM and SGD optimizers are used for training neural networks
- support for batch training
- parallel graph sampling is done using multiple threads
- negative graph sampling is a sampling where the final graph consists only of edges that don't exist
- evaluating the model's training performance using a variety of metrics like AUC, Precision, Recall, Accuracy, Confusion matrix
- evaluating the model's recommendation performance with Precision@k, Recall@k, F1@k and Average Precision metrics
If you want to try-out our implementation, head to github/memgraph/mage and find python/link_prediction.py. Feel free to give us a ⭐ if you like the code. The easiest way to test link-prediction is by downloading Memgraph Platform and using some of the preloaded datasets in Memgraph Lab.
There are some things you should be careful about when using link prediction:
- features of all nodes should be called the same (e.g saved as 'features' property in Memgraph)
- model's performance on the validation set is obtained using transductive splitting mode, while inductive dataset split is not yet supported. You can find more information about graph splitting on slides of Graph Machine Learning course offered by Stanford.
- to improve performance, self-loop is added to each node with the edge-type set to
self - the user can set the flag to automatically add reverse edges to each existing edge and hence, convert a directed graph to a bidirected one. If the source and destination nodes of the edge are the same, reverse edge type will be the same as the original edge type. Otherwise, the prefix rev_ will be added to the original edge type. See the FAQ part to further see why are self-loops and reverse edges very important in ML training and how you can get into problems if your graph is already undirected 🤔
Feel free to open a GitHub issue or start a discussion on Discord if you want to speed up development.
Usage
The following procedure is expected when using link prediction module:
- set parameters by calling
set_model_parametersfunction - train a model by calling
trainfunction - inspect training results (optional) by calling
get_training_resultsfunction - predict the relationship between two vertices by calling
predictor - call the
recommendfunction to find the most likely relationships
Implementation details
For the underlying GNN training we use the DGL library.
Fast and memory-efficient message passing primitives for training Graph Neural Networks. Scale to giant graphs via multi-GPU acceleration and distributed training infrastructure.
-- DGL team
Splitting the dataset
If the user specifies split_ratio 1.0, the model will train normally on a whole dataset without validating its performance on a validation set. However, if the user-defined split_ratio is a value between 0.0 and 1.0 but the graph is too small to have such a split, an exception will be thrown.
Self-loops
Self-loop edge is added to every node to improve link_prediction performance if specified by the user. Self-loop edges are added only as edge_type self, not in any other way, and to enable this, a custom module has been added.
Batch training
In heterogeneous graphs, all edges are used for creating the node’s neighbourhood but trained on only one edge type that can be set by the user.
For each gradient descent step, we select a mini-batch of nodes whose final representations at the L-th layer are to be computed. We then take all or some of their neighbours at the L−1 layer. This process continues until we reach the input. This iterative process builds the dependency graph starting from the output and working backwards to the input, as the figure below shows:
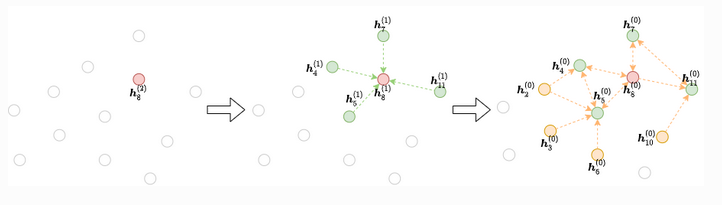
-- DGL docs
The reader is encouraged to take a look at the DGL mini-batch explanation for more details.
Procedures
If you want to execute this algorithm on graph projections, subgraphs or portions of the graph, be sure to check out the guide on How to run a MAGE module on subgraphs.
The link prediction module is organized as a stateful module in which the user can run several methods one after another without losing the context. The user should start with setting the parameters that are going to be used in the training. If the graph is heterogeneous (more than one edge type), target_relation parameter must be set so the model could distinguish supervision edges (edges used in prediction) from message passing edges (used for message aggregation). In the case of homogeneous graph, target relation will be automatically inferred. Node_features_property must also be sent by the user to specify where are saved original node features. Those are needed by graph neural networks to compute node embeddings. All other parameters are optional.
set_model_parameters()
Here is the description of all parameters supported by link prediction that you can set by calling the set_model_parameters method:
Input:
| Name | Type | Default | Description |
|---|---|---|---|
hidden_features_size | mgp.List[int] | [16, 16] | Defines the size of each hidden layer in the architecture. Input feature size is determined automatically while converting the original graph to the DGL compatible one. |
layer_type | str | graph_attn | Supported values are graph_sage and graph_attn. |
num_epochs | int | 100 | The number of epochs for model training. |
optimizer | str | ADAM | Supported values are ADAM and SGD. |
learning_rate | float | 0.01 | Optimizer's learning rate. |
split_ratio | float | 0.8 | The split ratio between the training and the validation set. There is no test dataset because it's assumed that the user first needs to create new edges in the original dataset to test a model on them. |
node_features_property | str | features | Property name where the node features are saved. |
device_type | str | cpu | Defines if the model will be trained using the CPU or Cuda GPU. To run on Cuda GPU, check if the system supports it with torch.cuda.is_available(), then set this flag to cuda. |
console_log_freq | int | 5 | Specifies how often results will be printed. This also directly specifies which results will be returned as training and validation results when calling the training method. |
checkpoint_freq | int | 5 | Select the number of epochs on which the model will be saved. The model is persisted on disc. |
aggregator | str | mean | Aggregator used in GraphSAGE model. Supported values are lstm, pool, mean and gcn. |
metrics | mgp.List[str] | [loss, accuracy, auc_score, precision, recall, f1, true_positives, true_negatives, false_positives, false_negatives] | Metrics used to evaluate the training model on the validation set. Additionally, epoch information will always be displayed. |
predictor_type | str | dot | Type of the predictor. A predictor is used for combining node scores to edge scores. Supported values are dot and mlp. |
attn_num_heads | List[int] | [4, 1] | GAT can support the usage of more than one head in each layer except the last one. The size of the list must be the same as the number of layers specified by the hidden_features_size parameter. |
tr_acc_patience | int | 8 | Training patience specifies for how many epochs drop in accuracy on the validation set is tolerated before the training is stopped. |
context_save_dir | str | None | Path where the model and predictor will be saved every checkpoint_freq epochs. |
target_relation | str | None | Unique edge type used for training. Users can provide only edge_type or tuple of the source node, edge type, dest_node if the same edge_type is used with more source-destination node combinations. |
num_neg_per_pos_edge | int | 1 | Number of negative edges that will be sampled per one positive edge in the mini-batch training. |
batch_size | int | 256 | Batch size used in both training and validation procedure. It specifies the number of indices in each batch. |
sampling_workers | int | 5 | Number of workers that will cooperate in the sampling procedure in the training and validation. |
last_activation_function | str | sigmoid | Activation function that is applied after the last layer in the model and before the predictor_type. Currently, only sigmoid is supported. |
add_reverse_edges | bool | False | Whether the module should add reverse edges for each existing edge in the obtained graph. If the source and destination node are of the same type, edges of the same edge type will be created. If the source and destination nodes are different, then the prefix rev_ will be added to the previous edge type. Reverse edges will be excluded as message passing edges for corresponding supervision edges. |
Output:
status: bool->Trueif all parameters were successfully updated,Falseotherwise.message: str->OKif all parameters were successfully updated,Error messageotherwise.
Only those parameters that need changing from their default values are sent when calling the procedure:
CALL link_prediction.set_model_parameters({num_epochs: 100, node_features_property: "features", tr_acc_patience: 8, target_relation: "CITES", batch_size: 256, last_activation_function: "sigmoid", add_reverse_edges: True})
YIELD status, message
RETURN status, message;
train()
The train method doesn't take any parameters, so it is very simple to use.
Output:
training_results: List[Dict[str, float]]-> List of training results through epochs. Model's performance is evaluated everyconsole_log_freqepochs.validation results: List[Dict[str, float]]-> List of validation results through epochs. Model's performance is evaluated everyconsole_log_freqepochs.
You can just call
CALL link_prediction.train()
YIELD training_results, validation_results
RETURN training_results, validation_results;
to get training and validation results summarized through epochs.
get_training_results()
The get_training_results method is used when the user wants to get performance data obtained from the last training. It is in the same form as a result of calling the training method. If there is no loaded model, the exception will be thrown.
CALL link_prediction.get_training_results()
YIELD training_results, validation_results;
RETURN training_results, validation_results;
Output:
training_results: List[Dict[str, float]]-> List of training results through epochs. Model's performance is evaluated everyconsole_log_freqepochs.validation results: List[Dict[str, float]]-> List of validation results through epochs. Model's performance is evaluated everyconsole_log_freqepochs.
predict()
The predict method takes two arguments, src_vertex and dest_vertex, and predicts whether there is an edge between them or not. It supports an “actual” prediction scenario when the edge doesn’t exist and the user wants to predict whether there is an edge or not but also a scenario in which there is an edge between two vertices and the user wants to check the model’s evaluation.
Input
src_vertex: mgp.Vertex-> Source vertex of the edgedest_vertex: mgp.Vertex-> Destination vertex of the edge.
Output
score: mgp.Number-> Score between 0 and 1 that represents the probability of two nodes being connected.
MATCH (v1:PAPER {id: "ID_1"})
MATCH (v2:PAPER {id: "ID_2"})
CALL link_prediction.predict(v1, v2)
YIELD score
RETURN score;
recommend()
The recommend method can be used to recommend the best k nodes from dest_vertices to src_vertex. It is implemented efficiently using the max heap data structure. The best nodes are determined based on the edge scores. Metrics specific to recommendation systems (precision@k, recall@k, f1@k and average precision) are logged to the standard output. K is equal to the given min(k, length(dest_vertices), length(results)) where results are a list of all recommendations given by the model(classified as a positive example.)
Input
src_vertex: mgp.Vertex→ Source node.dest_vertices: List[mgp.Vertex]→ destination nodes. If they are not of the same type, an exception is thrown.k: int→ Number of edges to recommend.
Output
score: mgp.Number→ Score between 0 and 1 that represents the probability of two nodes being connected.recommendation: mgp.Vertex→ A reference to the target node.
MATCH (v1:Customer {id: "8779-QRDMV"})
MATCH (p:Plan)
WITH collect(p) AS all_plans, v1
CALL link_prediction.recommend(v1, all_plans, 5)
YIELD score, recommendation
RETURN v1, score, recommendation;
load_context()
Loading the context means loading the model and the predictor. If the user specifies the path, the method will try to load it from there. Otherwise, context will be loaded from the default parameter specified in the link_prediction_parameters module.
Input
path: str→ Path to the folder where the model and the predictor are saved.
Output
status: mgp.Any→ True to indicate that execution went well.
CALL link_prediction.load_context() YIELD * RETURN *;
reset_parameters()
You can explicitly reset parameters whenever you want. Note, however, that parameters will be reset before the training even if not specified because of implementation reasons.
Output
status: mgp.Any→ True to indicate that method is successfully finished.
CALL link_prediction.reset_parameters() YIELD * RETURN *;
Results
We extensively tested our model on the CORA dataset and the Telecom recommendation dataset. To show you how the training performance could progress through epochs, here are the results for one of our basic models tried on the Cora dataset:
| epoch_num | AUC | accuracy | precision | recall | f1 |
|---|---|---|---|---|---|
| 1 | 0.64 | 0.594 | 0.613 | 0.494 | 0.547 |
| 2 | 0.781 | 0.696 | 0.711 | 0.663 | 0.686 |
| 3 | 0.798 | 0.729 | 0.752 | 0.682 | 0.715 |
| 4 | 0.754 | 0.686 | 0.716 | 0.617 | 0.663 |
| 5 | 0.789 | 0.711 | 0.715 | 0.7 | 0.707 |
| 6 | 0.813 | 0.756 | 0.742 | 0.784 | 0.763 |
| 7 | 0.884 | 0.772 | 0.764 | 0.791 | 0.775 |
| 8 | 0.859 | 0.775 | 0.781 | 0.766 | 0.773 |
| 9 | 0.871 | 0.805 | 0.822 | 0.777 | 0.798 |
| 10 | 0.832 | 0.759 | 0.776 | 0.729 | 0.752 |
Example
- Step 1: Input graph
- Step 2: Load commands
- Step 3: Set model parameters
- Step 4: Train
- Step 5: Train results
- Step 6: Predict
- Step 7: Predict results
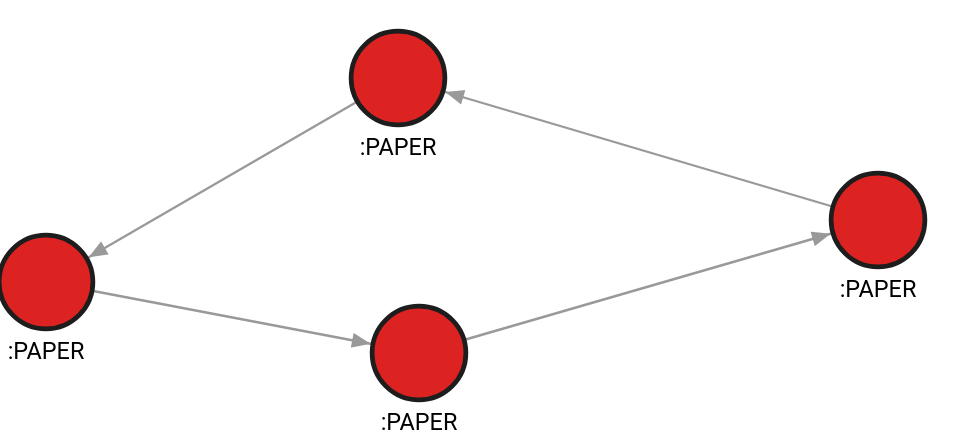
CREATE (v1:PAPER {id: 10, features: [1, 2, 3]});
CREATE (v2:PAPER {id: 11, features: [1.54, 0.3, 1.78]});
CREATE (v3:PAPER {id: 12, features: [0.5, 1, 4.5]});
CREATE (v4:PAPER {id: 13, features: [0.78, 0.234, 1.2]});
MATCH (v1:PAPER {id: 10}), (v2:PAPER {id: 11}) CREATE (v1)-[e:CITES {}]->(v2);
MATCH (v2:PAPER {id: 11}), (v3:PAPER {id: 12}) CREATE (v2)-[e:CITES {}]->(v3);
MATCH (v3:PAPER {id: 12}), (v4:PAPER {id: 13}) CREATE (v3)-[e:CITES {}]->(v4);
MATCH (v4:PAPER {id: 13}), (v1:PAPER {id: 10}) CREATE (v4)-[e:CITES {}]->(v1);
CALL link_prediction.set_model_parameters({target_relation: ["PAPER", "CITES", "PAPER"], node_features_property: "features",
split_ratio: 1.0, predictor_type: "mlp", num_epochs: 100, hidden_features_size: [256], attn_num_heads: [1]}) YIELD * RETURN *;
CALL link_prediction.train() YIELD training_results, validation_results
RETURN training_results, validation_results;
+--------------------+--------------------+--------------------+--------------------+--------------------+
| epoch_num | accuracy | auc_score | loss | precision |
+--------------------+--------------------+--------------------+--------------------+--------------------+
| 17 | 0.833 | 0.906 | 0.428 | 1.0 |
| 18 | 0.917 | 0.938 | 0.393 | 1.0 |
| 19 | 0.833 | 0.938 | 0.365 | 0.75 |
| 20 | 0.917 | 0.938 | 0.341 | 1.0 |
| 21 | 0.917 | 0.938 | 0.315 | 1.0 |
| 22 | 0.833 | 0.969 | 0.296 | 0.75 |
| 23 | 0.917 | 1.0 | 0.277 | 1.0 |
| 24 | 0.917 | 1.0 | 0.246 | 0.8 |
| 25 | 0.917 | 1.0 | 0.233 | 0.8 |
| 26 | 1.0 | 1.0 | 0.202 | 1.0 |
MATCH (v1:PAPER {id: 10})
MATCH (v2:PAPER {id: 12})
CALL link_prediction.predict(v1, v2)
YIELD score
RETURN score;
+-------+
| score |
| 0.104 |
FAQ
Why can I get into problems with reverse edges?
Having a reverse_edge in your dataset can be a problem if they are not excluded from message passing edges in the prediction of its opposite edge(supervision edge). The best thing you can do is have a directed graph and the module will automatically add reverse edges, if you specify add_reverse_edges in the set_model_parameters method, in a way that doesn't cause information flow.
What is a transductive dataset split?
The transductive dataset split assumes that the entire graph can be observed in all dataset splits. We distinguish four types of edges, and those are: validation, training, message passing and supervision edges.
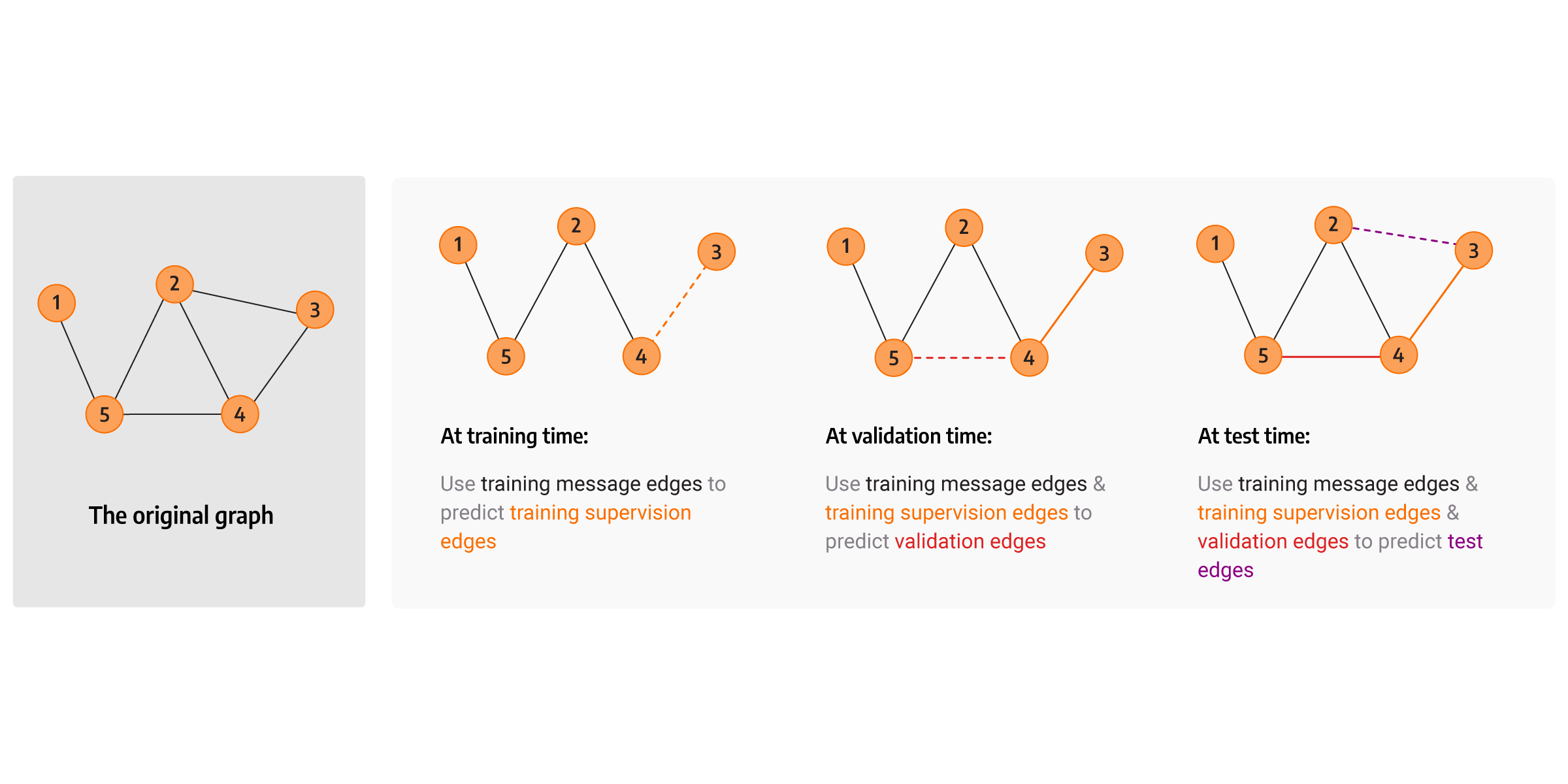
The transductive dataset split is described in detail by prof. Jure Leskovec at one of its presentations for Graph ML course.
