Node assortativity coefficients and correlation measures#
In this tutorial, we will go through the theory of assortativity and its measures.
Specifically, we’ll focus on assortativity measures available in NetworkX at algorithms/assortativity/correlation.py:
Attribute assortativity
Numeric assortativity
Degree assortativity
as well as mixing matrices, which are closely releated to assortativity measures.
Assortativity#
Assortativity in a network refers to the tendency of nodes to connect with other ‘similar’ nodes over ‘dissimilar’ nodes.
Here we say that two nodes are ‘similar’ with respect to a property if they have the same value of that property. Properties can be any structural properties like the degree of a node to other properties like weight, or capacity.
Based on these properties we can have a different measure of assortativity for the network. On the other hand, we can also have disassortativity, in which case nodes tend to connect to dissimilar nodes over similar nodes.
Assortativity coefficients#
Let’s say we have a network
Mixing matrix#
Let the property
Now define,
in Python code it would look something like a = e.sum(axis=0) and b = e.sum(axis=1)
Finally, let
Then we can define the assortativity coefficient for this property based on the Pearson correlation coefficient.
Attribute Assortativity Coefficient#
Here the property
From here onwards we will use subscript notation to denote indexing, for eg.
It is implemented as attribute_assortativity_coefficient.
Numeric Assortativity Coefficient#
Here the property
It is implemented as numeric_assortativity_coefficient.
Degree Assortativity Coefficient#
When it comes to measuring degree assortativity for directed networks we have
more options compared to assortativity w.r.t a property because we have 2 types
of degrees, namely in-degree and out-degree.
Based on the 2 types of degrees we can measure
r(in,in) : Measures tendency of having a directed edge (u,v) such that, in-degree(u) = in-degree(v).
r(in,out) : Measures tendency of having a directed edge (u,v) such that, in-degree(u) = out-degree(v).
r(out,in) : Measures tendency of having a directed edge (u,v) such that, out-degree(u) = in-degree(v).
r(out,out) : Measures tendency of having a directed edge (u,v) such that, out-degree(u) = out-degree(v).
Note: If the network is undirected all the 4 types of degree assortativity are the same.
To define the degree assortativity coefficient for all 4 types we need slight
modification in the definition of
Let
It is implemented as degree_assortativity_coefficient and
degree_pearson_correlation_coefficient. The latter function uses
scipy.stats.pearsonr to calculate the assortativity coefficient which makes
it potentally faster.
Example#
%matplotlib inline
import networkx as nx
import matplotlib.pyplot as plt
import pickle
import copy
import random
import warnings
warnings.filterwarnings("ignore")
Illustrating how value of assortativity changes
gname = "g2"
# loading the graph
G = nx.read_graphml(f"data/{gname}.graphml")
with open(f"data/pos_{gname}", "rb") as fp:
pos = pickle.load(fp)
fig, axes = plt.subplots(4, 2, figsize=(20, 20))
# assign colors and labels to nodes based on their 'cluster' and 'num_prop' property
node_colors = ["orange" if G.nodes[u]["cluster"] == "K5" else "cyan" for u in G.nodes]
node_labels = {u: G.nodes[u]["num_prop"] for u in G.nodes}
for i in range(8):
g = nx.read_graphml(f"data/{gname}_{i}.graphml")
# calculating the assortativity coefficients wrt different proeprties
cr = nx.attribute_assortativity_coefficient(g, "cluster")
r_in_out = nx.degree_assortativity_coefficient(g, x="in", y="out")
nr = nx.numeric_assortativity_coefficient(g, "num_prop")
# drawing the network
nx.draw_networkx_nodes(
g, pos=pos, node_size=300, ax=axes[i // 2][i % 2], node_color=node_colors
)
nx.draw_networkx_labels(g, pos=pos, labels=node_labels, ax=axes[i // 2][i % 2])
nx.draw_networkx_edges(g, pos=pos, ax=axes[i // 2][i % 2], edge_color="0.7")
axes[i // 2][i % 2].set_title(
f"Attribute assortativity coefficient = {cr:.3}\nNumeric assortativity coefficient = {nr:.3}\nr(in,out) = {r_in_out:.3}",
size=15,
)
fig.tight_layout()
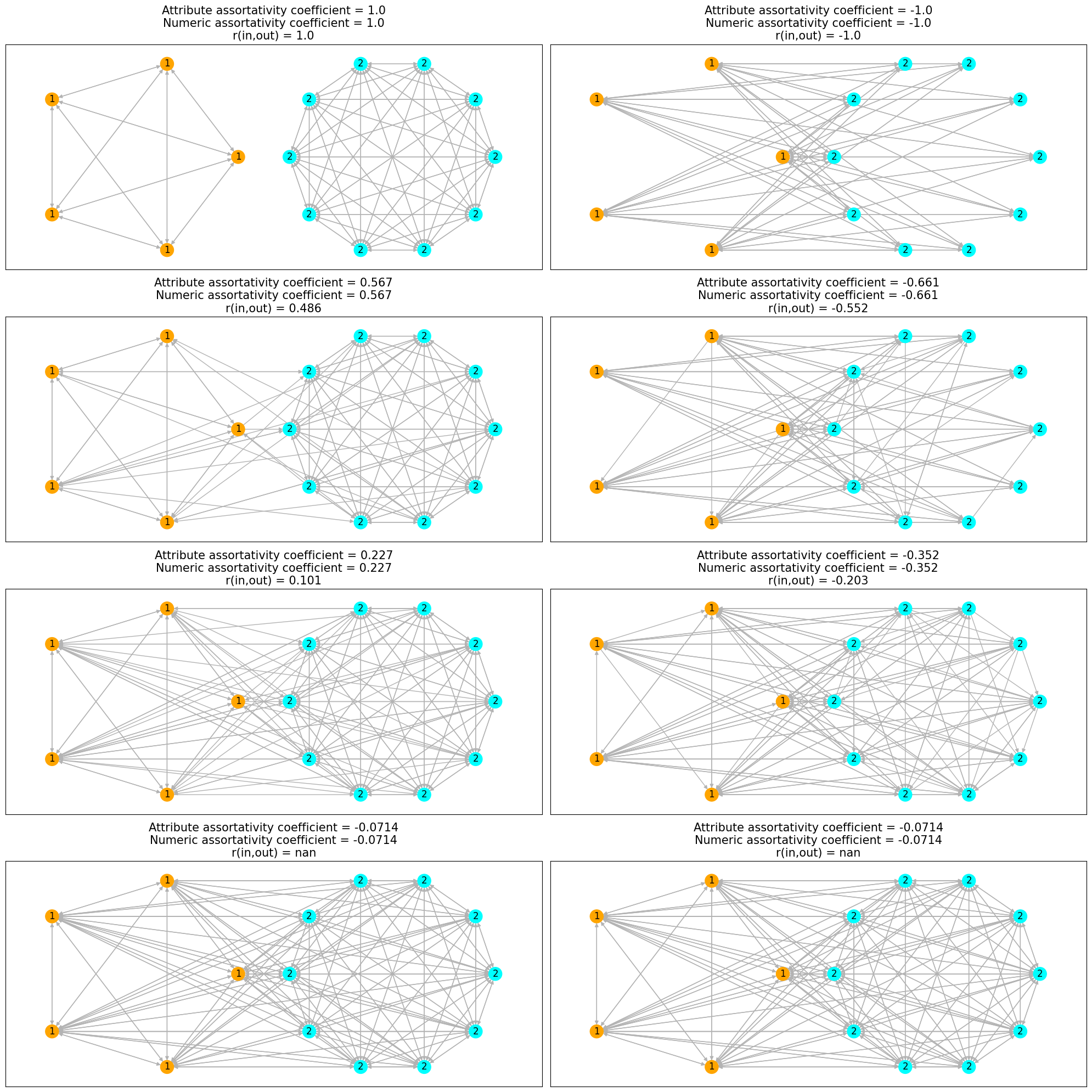
Nodes are colored by the cluster property and labeled by num_prop property.
We can observe that the initial network on the left side is completely assortative
and its complement on right side is completely disassortative.
As we add edges between nodes of different (similar) attributes in the assortative
(disassortative) network, the network tends to a non-assortative network and
value of both the assortativity coefficients tends to
The parameter nodes in attribute_assortativity_coefficient and
numeric_assortativity_coefficient specifies the nodes whose edges are to be
considered in the mixing matrix calculation.
That is to say, if nodes.
For the undirected case, it’s considered if atleast one of the nodes.
The nodes parameter is interpreted differently in degree_assortativity_coefficient and
degree_pearson_correlation_coefficient, where it specifies the nodes forming a subgraph
whose edges are considered in the mixing matrix calculation.
# list of nodes to consider for the i'th network in the example
# Note: passing 'None' means to consider all the nodes
nodes_list = [
None,
[str(i) for i in range(3)],
[str(i) for i in range(4)],
[str(i) for i in range(5)],
[str(i) for i in range(4, 8)],
[str(i) for i in range(5, 10)],
]
fig, axes = plt.subplots(3, 2, figsize=(20, 16))
def color_node(u, nodes):
"""Utility function to give the color of a node based on its attribute"""
if u not in nodes:
return "0.85"
if G.nodes[u]["cluster"] == "K5":
return "orange"
else:
return "cyan"
# adding a edge to show edge cases
G.add_edge("4", "5")
for nodes, ax in zip(nodes_list, axes.ravel()):
# calculating the value of assortativity
cr = nx.attribute_assortativity_coefficient(G, "cluster", nodes=nodes)
nr = nx.numeric_assortativity_coefficient(G, "num_prop", nodes=nodes)
# drawing network
ax.set_title(
f"Attribute assortativity coefficient: {cr:.3}\nNumeric assortativity coefficient: {nr:.3}\nNodes = {nodes}",
size=15,
)
if nodes is None:
nodes = [u for u in G.nodes()]
node_colors = [color_node(u, nodes) for u in G.nodes]
nx.draw_networkx_nodes(G, pos=pos, node_size=450, ax=ax, node_color=node_colors)
nx.draw_networkx_labels(G, pos, labels={u: u for u in G.nodes}, font_size=15, ax=ax)
nx.draw_networkx_edges(
G,
pos=pos,
edgelist=[(u, v) for u, v in G.edges if u in nodes],
ax=ax,
edge_color="0.3",
)
fig.tight_layout()
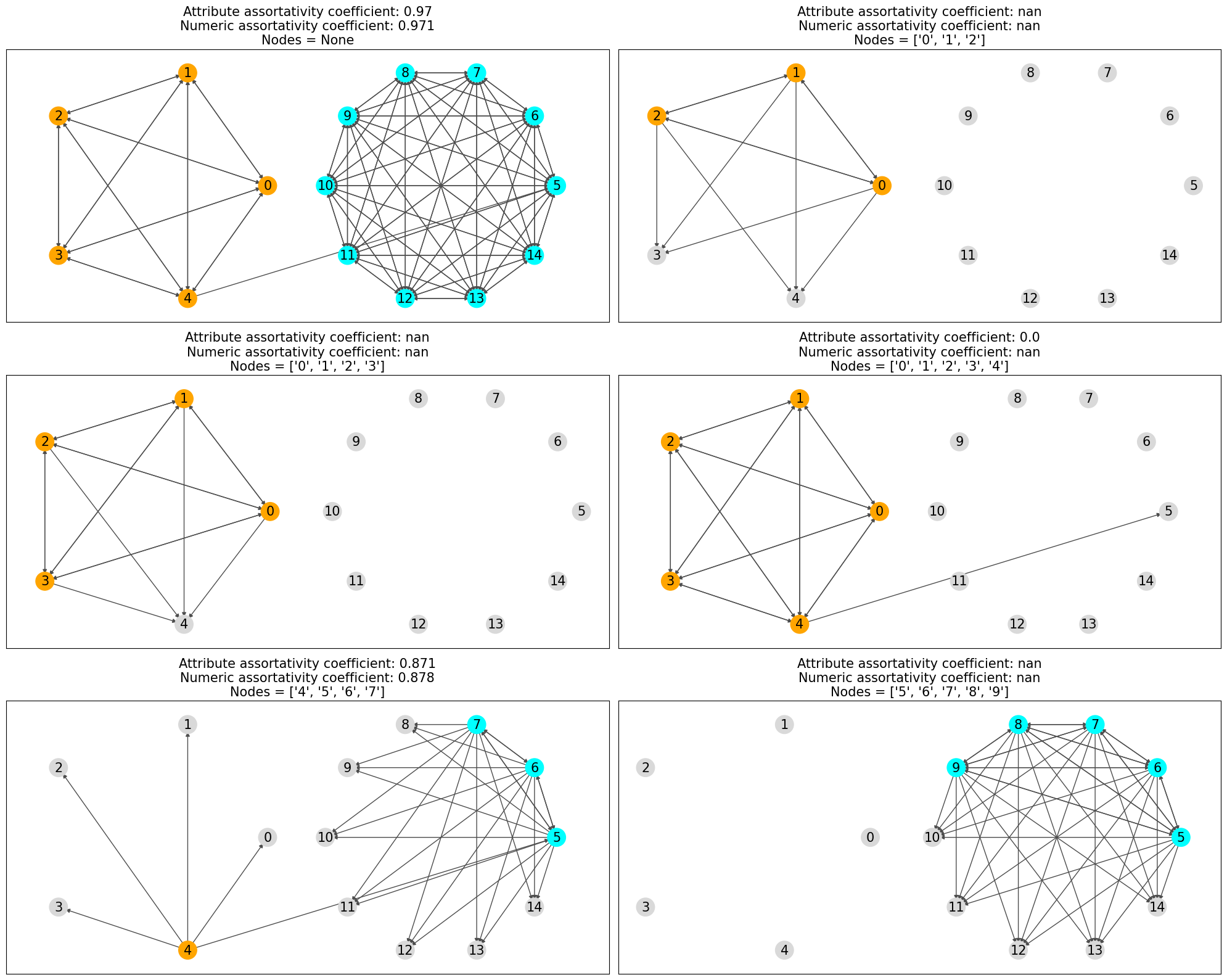
In the above plots only the nodes which are considered are colored and rest are grayed out and only the edges which are considerd in the assortaivty calculation are drawn.
